Meet our team
We are an enthusiastic bunch of researchers working on AI/ML applications in the life sciences.

Oliver Stegle
Director
DKFZ
EMBL
Our interest lies in computational approaches for unravelling molecular and phenotypic variation. How do genetic background and environment jointly shape phenotypic traits or cause diseases? How are genetic and external factors integrated at different molecular levels, and how variable are these molecular readouts between individual cells?
We use statistical inference and machine learning as our main tools to address these questions. The methods we develop allow for exploiting large and high-dimensional omics data to identify disease signatures and pinpoint causal drivers. Our laboratory combines principles from classical statistics, machine learning and causal reasoning. Current research aims to include improved association tests for genome-wide association studies (GWAS) that scale to millions of samples while accounting for technical and biological confounding factors. A second aim is dimensionality reduction that allows for integrating data across omics modalities, profiled across space and time.
Our methodological research aims to tie in with experimental collaborations, in the context of which we develop methods to fully exploit large-scale datasets obtained using the most recent profiling technologies. With the help of our international collaborations and networks we have established population-scale genetic and molecular data resources that allow for unravelling gene regulatory dependencies with unprecedented resolution in pluripotent cells and in cancer. We are also actively advancing multi-omics profiling methods in single cells and spatial omics applied to a variety of questions in basic biology as well as in disease contexts.
The ability to fully exploit novel algorithms and machine learning, access to large and well-integrated datasets is indispensable. To this end, we are coordinating the German Human Genome-Phenome Archive (GHGA), a major national network that seeks to bring together omics variation data across Germany and Europe, and will deliver key opportunities for federated learning and access to large patient datasets in the future.

Anna Kreshuk
Co-director
EMBL
We are a part of the Cell Biology and Biophysics Unit at EMBL. In addition to basic biology research, our colleagues at the Unit develop novel microscopy technologies and theoretical approaches to the interpretation of biological images. Our research is also concerned with microscopy, most broadly with the extraction of objects of biological interest from large, volumetric, challenging datasets. For example, we are interested in semantic and instance segmentation problems in the context of biology, as well as in computational image reconstruction for novel microscopy modalities. Besides methods development, we are very invested in the cause of accessible end-user software, bringing machine learning-based image analysis methods to life scientists without computational expertise. Our group of computer scientists and physicists works in close collaboration with biologists and microscopists, providing - in addition to more generic algorithms - solutions to outstanding real-world problems of cutting-edge life sciences research.
Previous and current research
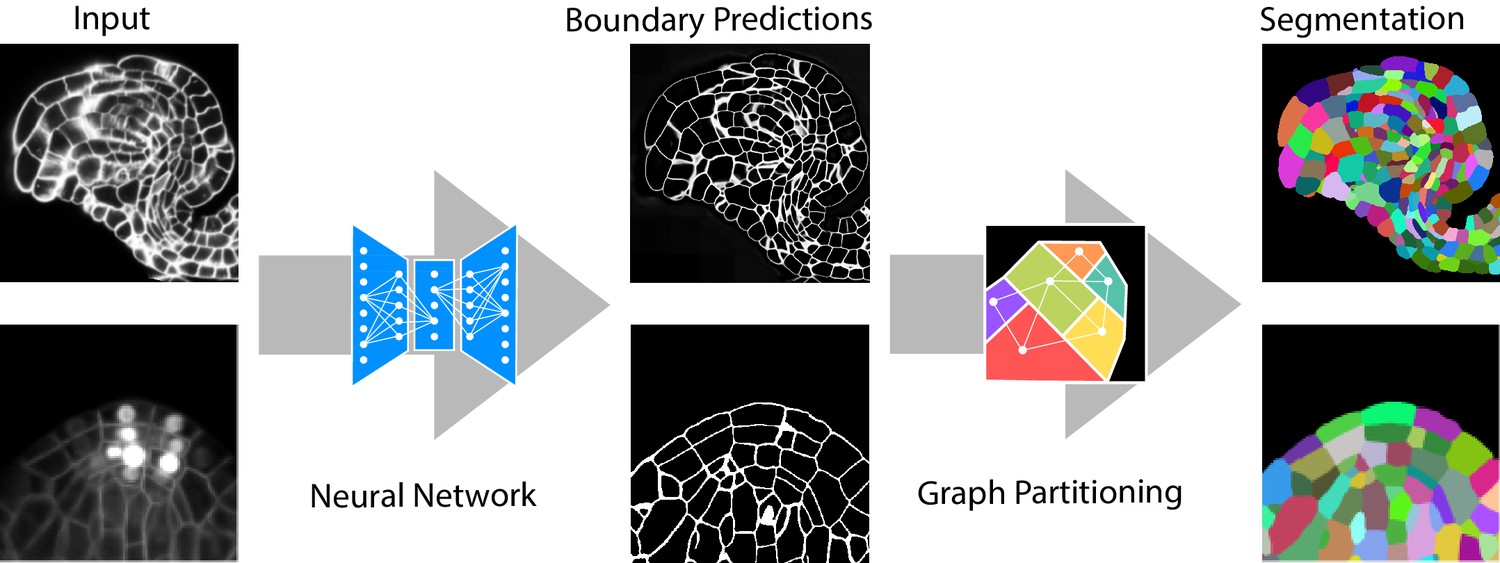
Our group is particularly interested in solving challenging segmentation problems for light or electron microscopy (LM or EM), in 3D and at large scale. Most recently, we have developed methods and tools to segment all cells and nuclei in a juvenile worm of the species Platynereis dumerilii (EM, Vergara et al., Cell 2021), as well as in various plant organs and tissues (LM, Wolny, Cerrone et al., eLife 2020). Here, we combined bottom-up deep learning approaches with top-down biological priors to achieve very accurate segmentations also for volumes of high intrinsic variability, in presence of substantial noise and with limited training data.
One of the major advantages of learning-based computer vision approaches is their general applicability: tailoring to a particular problem is performed by providing suitable training data, while the core of the algorithm remains unchanged. To bring these methods to members of the life science community without computer vision expertise, we have developed a toolkit for interactive learning and segmentation (ilastik). The development of ilastik started in Fred Hamprecht’s group at the University of Heidelberg and now continues with us at EMBL, providing an easy gateway to machine learning for tens of thousands of users.
Future projects and goals
All machine learning algorithms require user guidance at the training stage, but deep learning – the driver of the current computer vision revolution – is even more annotation hungry. This problem is especially acute in biological imaging, where annotation of ground-truth data cannot easily be outsourced to non-experts, and changes in experimental conditions often require retraining. Besides the annotation burden, the training process itself depends upon non-trivial expertise in the choice and tuning of hyperparameters. Our group is currently working on methods and training strategies that would reduce the requirements on the amount of training data, which we hope to incorporate into ilastik (Wolny et al., arxiv, 2021). We are also interested in creative combinations of deep learning and microscopy (Wagner, Beuttenmuelluer et al., Nature Methods 2021) and learning-based analysis of morphology.

Carsten Rother
Co-director
Heidelberg University
We are the “Computer Vision and Learning Lab” at Heidelberg University and I run the “3D Computer Vision” group within the lab. Computer Vision aims at extracting high-level information from images and videos, such as reconstructing a 3-dimensional scene, detecting and tracking objects in video, or synthesizing new images. In my group we focus on tasks that reason about the 3-dimensional nature of the world that was captured. Examples include tracking of a dynamic face in 3D or inserting a 3D object into existing footage – in order to generate training data for deep learning. To solve these tasks, we develop new Machine Learning methods, where often the best strategy is to bake in geometric knowledge and algorithms into new trainable neural networks. Apart from the methodological aspects, I am also interested in developing solutions that are beneficial to society. One example is a novel solution for multi-party video conferencing where we collaborate with researchers from other disciplines, such as Human-Computer Interaction. To this end, my group has a history of creating start-ups based on our research.

Björn Ommer
Heidelberg University
The Ommer lab at the Interdisciplinary Center for Scientific Computing (IWR) conducts research in Computer Vision and Machine Learning. In particular, we are interested in all aspects of scene understanding, visual synthesis, deep metric and representation learning, and image retrieval. Since annotation of training data is costly, we especially develop self-supervised and weakly supervised approaches to learning. Moreover, the inferred models should not only be powerful representations of our world, but they should also be interpretable by a user. Rather than presenting the user with a black box, we consequently seek an interpretable model that draws conclusions from the input data. We have been developing these methods in the context of long-term interdisciplinary collaborations. Within the life sciences we are particularly interested in non-invasive diagnostic tools for the neurosciences that can be efficiently integrated into clinical or scientific practice since they require little to no supervision by the expert users.
Visual Synthesis and Interpretable AI with Disentangled Representations
Deep learning has significantly improved the expressiveness of representations. However, present research still fails to understand why and how they work and cannot reliably predict when they fail. Moreover, the different characteristics of our physical world are commonly intermingled, making it impossible to study them individually. We incorporate novel paradigms for disentangling multiple object characteristics and present interpretable models to translate arbitrary network representations into semantically meaningful, interpretable concepts. We also obtain disentangled generative models that explain their latent representations by synthesis while being able to alter different object characteristics individually.
Deep Metric and Representation Learning
To understand visual content, computers need to learn what makes images similar. This similarity learning directly implies a representation of the visual content that captures the inherent structure of the data. We present several approaches that can be applied on top of arbitrary deep metric learning methods and various network architectures. Key issues that these works tackle include improving generalization and transfer to novel data, shared feature learning, and adaptive sampling strategies based on reinforcement learning to effectively utilize large amounts of training data.

Fred A. Hamprecht
Heidelberg University
We develop machine learning algorithms for image analysis. More specifically, we are interested in principled methods that ingest images or video, and output “structured” predictions, such as the partitioning of an image into its constituent parts, or the tracking of all objects in a video. Most of our applications are in the life sciences; for instance, we segment all cells in a brain or a plant, or track virus components before assembly.
We focus on methods that have a sound mathematical background, such as combinatorial optimization or algebraic graph theory, while being widely applicable and useful in practice. We attach particular importance to methods that can be trained with few examples, or by weak supervision, to empower colleagues from other disciplines to analyze their own data, conveniently. To the same end, we have launched the ilastik program for interactive machine learning that is now headed by Anna Kreshuk.
Personally, I enjoy and feel most privileged to be able to work on things unknown, and to teach the next generation of scientists and engineers. I am proud that several past PhD students continue to serve in research and education, including:
Anna Kreshuk, Group Leader, EMBL
Bjoern Andres, Professor, TU Dresden
Bjoern Menze, Professor, TU München
Bernhard Renard, Professor, HPI, Potsdam University
Luckily, I am blessed with a fantastic team of researchers who happen to be both extremely gifted and nice.

Jan Korbel
EMBL
The Korbel group combines computational and experimental approaches, including in single cells, to unravel determinants and consequences of germline and somatic genetic variation with a special focus on disease mechanisms.
Genetic variation studies have uncovered that genomic structural variants (SVs) such as deletions, insertions, and inversions account for most varying bases in human genomes. Recent studies indicate that somatic SVs occur post-zygotically throughout our lifespan, and show association with ageing and human diseases - calling into question the long-held belief that the genome is largely static within an individual, and preserved across all cells therein.
We employ a diversity of omics and imaging approaches - from single-cell multi-omics to spatial and bulk-cell omics as well as state-of-the-art microscopy - to investigate molecular mechanisms behind complex human phenotypes associated with genetic variants.
In addition to experimental methodologies applied to tissues and organoids, our laboratory is devising data science techniques including state-of-the-art machine learning methods for processing high-dimensional single-cell data sets, and for coupling genetic variation discovery with molecular and clinical phenotype data.
Previous and current research
Our particular interest lies in understanding patterns of genetic mosaicism at cellular resolution. Our scTRIP method (Sanders et al., Nat Biotechnol 2020, Fig. 1) enables the direct detection of SV mutational processes in single cells, and as such can be used to obtain insights into pathomechanisms acting in human tissues.
Another interest centres around uncovering commonalities and differences between molecular disease mechanisms in disparate cancer entities. In a rare-variant association study in medulloblastoma (MB) genomes/exomes, we recently described rare germline loss-of-function variants in the Elongator complex protein 1 (ELP1) gene in 15% of childhood MB genomes driven by Sonic hedgehog signalling (Waszak et al., Nature 2020). ELP1-associated MBs exhibit somatic loss of the wild-type ELP1 allele mediated by somatic large deletions that concomitantly cause loss of the PTCH1 gene residing adjacent to ELP1 on chromosome 9, involving an intriguing ‘three-hit’ molecular process (Fig. 2).
With respect to data science, our group has pioneered the utilisation of cloud computing to enable the global sharing and processing of large-scale biological data. We co-initiated and co-led the ICGC/TCGA Pan-Cancer Analysis of Whole Genomes (PCAWG) project, an international study for sharing cancer genomes. Our group is also actively involved in building the German Human Genome-Phenome Archive (GHGA), a national research data infrastructure for disseminating and federating human genomics data from German studies nationally and internationally.

Julio Saez-Rodriguez
Heidelberg University
Our major interest is the use of machine learning in combination with biological knowledge to analyze big molecular data to better understand and treat disease. We believe that this biological knowledge can be instrumental to move from pure correlation to causation in large data sets, and thereby identify the molecular processes that underlie specific phenomena. Besides developing and applying methods that we share as free open-source packages, we are also involved in the benchmarking of methods, in our own research but also as a crowdsourcing as part of the DREAM challenges.
Our current focus is on the analysis of transcriptomic, metabolomic, and proteomic data sets, increasingly with resolution at the single-cell and spatial level. These data sets come from laboratory experiments as well as from patients suffering an array of conditions ranging from cancer to kidney and cardiovascular disease. We integrate the pertinent biologica knowledge in our meta-resource OmniPath. We use these data sets and knowledge to build models that are both mechanistic (to provide understanding) and predictive (to generate novel hypotheses). These models, in turn, we use to find disease biomarkers and improved therapeutic opportunities.

Klaus H. Maier-Hein
DKFZ
Our ambition is to improve the ability of machines to analyze and interpret medical image data with a focus on generalizability and uncertainty.
For this purpose our work is focused on deep learning methodology in the context of medical image analysis, with various clinical applications that we are working on together with radiologists, oncologists and radiation therapists. We are also interested in the concept of federated learning and are therefore developing the required methods and the research software infrastructure needed for multi-site projects.

Lena Maier-Hein
DKFZ
Medical interventions are among the fundamental pillars of healthcare. Our mission at the Division of Computer Assisted Medical Interventions is to improve the quality of interventional healthcare in a data-driven manner. To this end, our multidisciplinary group builds upon principles and knowledge from a diversity of research fields including artificial intelligence (AI), statistics, computer vision, biophotonics and medicine. We are committed to the ultimate goal of creating benefits for patients and medical staff, and we aim to develop a holistic concept spanning the three significant topics - perception, data interpretation and real-time assistance (see Figure) - and connecting them through a cycle of continuous learning. Novel spectral imaging techniques enabled by deep learning are being developed as safe, reliable and real-time imaging modalities during interventions. When interpreting the perceived data in the context of available knowledge, we specifically address common roadblocks to clinical translation such as data sparsity, explainability and uncertainty handling. In close collaboration with our clinical partners, we leverage these methods for the development of context-aware interventional assistance systems. Finally, we place a strong focus on the reliable validation of AI algorithms for clinical purposes.

Ullrich Köthe
Heidelberg University
We make up the “Explainable Machine Learning” group in the Computer Vision and Learning Lab. Our work focuses on deep learning methodology that strives to establish machine learning as a new way to gain insight and create new knowledge in the sciences. Models that merely make somewhat accurate predictions in a blackbox manner are not good enough for this goal. We therefore investigate the design and theoretical foundations of novel network architectures and training algorithms, which provide reliable self-assessment of their uncertainty, emerge humanly interpretable latent representations and are robust against distribution shifts in the data. In particular, we utilise invertible neural networks as an especially promising approach to the efficient and well-founded probabilistic treatment of inverse problems and generative modeling. We have demonstrated the utility of these solutions in numerous applications from physics and astronomy to computer vision, biology, and medicine. To achieve our goals, we lay emphasis on interdisciplinary work with other fields of science, and on the rapid dissemination of its results via reusable open-source software libraries.

Wolfgang Huber
EMBL
Biology is now a science of big data, but the data are often heterogeneous, noisy, incomplete, replete with biases, poorly calibrated (“batch effects”), and indirect—what is being measured and its resolution is contingent on current technology rather than what would be desirable. With our work, we try to address these challenges, driven by the motivation to better empower machine learning and statistics for biotechnological experimentation and observation, and for scientific discovery in biology and biomedicine.
We aim to develop and improve new data generating technologies in biological research, such as single cell omics, imaging, mass spectrometry, high-throughput genetic and chemical perturbation assays, by powering them with the best statistical and machine learning methods. This includes inference—reasoning with uncertainty, providing optimal estimates or making optimal decisions based on incomplete, noisy or overwhelming data—as well as data exploration, visualization and discovery: helping scientists examine large, complex datasets that they could not grasp otherwise.
We are interested in new datasets of unprecedented scope and quality. To this end, we work with biomedical researchers who are assembling such experiments and studies, and together we optimize all phases from experiment/study design, data management, quality control, to exploitation.
We believe that good science is open science, and we consider the release and maintenance of re-usable, interoperable, collaborative scientific software an integral part of computational and statistical method development, and of scientific publishing more generally. We contribute to the Bioconductor Project, an open source software collaboration to provide tools for the analysis and study of high-throughput genomic data.
Together with Susan Holmes, Huber co-authored the textbook “Modern Statistics for Modern Biology” (Cambridge University Press, ISBN 9781108705295)